CLI & C++ User Manual
This page contains an in-depth introduction to Yggdrasil Decision Forests (YDF) CLI API. The content presented on this page is generally not necessary to use YDF, but it will help users improve their understanding and use advance options.
New users should first check out the Quick start.
Most concepts presented here apply to the other APIs, notably the C++ API.
Dataset
A dataset is a list of examples. Datasets can be stored in memory, on disk, or generated on the fly. Examples are ordered in a dataset, that is an example in a dataset is unambiguously identified by its index in a dataset.
An attribute (also known as a feature, variable, or column in other libraries) refers to one particular piece of information in a tabular dataset. For example, the age, name and country of birth of a person are attributes.
Each attribute has a semantic (also known as type). The semantic determines how an attribute is used by a model. For example, age (e.g., number of years since birth) is generally numerical while country (e.g., ISO 3166 country code) is categorical.
YDF supports the following semantics.
-
NUMERICAL: Numerical value. Generally for quantities or counts with full ordering. For example, the age of a person, or the number of items in a bag. Can be a float or an integer. Missing values are represented with NaN.
-
CATEGORICAL: A categorical value. Generally for a type/class with a finite set of possible values without ordering. For example, the color RED in the set {RED, BLUE, GREEN}. Can be a string or an integer. Internally, categorical values are stored as int32 and should therefore be smaller than ~2B.
-
CATEGORICAL_SET: Set of categorical values. Great to represent tokenized texts. Unlike CATEGORICAL, the number of items in a CATEGORICAL_SET can change and the order/index of each item doesn't matter.
-
BOOLEAN: Boolean value. Similar to CATEGORICAL, but with only two possible values.
-
DISCRETIZED_NUMERICAL: Numerical values automatically discretized into bins. Discretized numerical features are faster to train than (non-discretized) numerical features. If the number of unique values of these features is lower than the number of bins, the discretization is lossless from the point of view of the model. If the number of unique values of this feature is greater than the number of bins, the discretization is lossy from the point of view of the model. Lossy discretization can reduce and sometimes increase (due to regularization) the quality of the model.
-
HASH: The hash of a string value. Used when only the equality between values is important (not the value itself). Currently, only used for groups in ranking problems e.g. the query in a query/document problem. The hashing is computed with farmhash.
An example is a collection of attribute values. Following is an example with three attributes:
"attribute_1": 5.1 # NUMERICAL
"attribute_2": "CAT" # CATEGORICAL stored as string
"attribute_3": [1, 2, 3] # CATEGORICAL-SET stored as integer
A data specification (or dataspec for short) is the definition of a list
of attributes. This definition contains the name, semantic and meta-data of each
attribute in the dataset. A dataspec is stored as a
ProtoBuffer (see its
definition at dataset/data_spec.proto). A dataspec can be printed in a
readable way using the show_dataspec CLI command.
Google ProtoBuffers are used extensively for configuration in YDF. Like XML or JSON, Protobuf is a language independent serialization format to exchange data. Protobuf can be in text of binary format. YDF configuration are always files containing protobuf V2 in text format.
A protobuf is always attached to a Protobuf definition that list the names and types of the allowed fields. In this documentation, several examples contain Protobuf text and a link to the corresponding Protobuf definition.
Following is an example of protobuf and corresponding protobuf definition:
Protobuf
Protobuf definition
message MyMessage{
optional int32 field_a = 1 [default = 5];
optional float field_b = 2 [default = 8.0];
}
In the definition, each field is assigned to a unique integer id in the form
<field name> = <field id> [default= <default value>].
This is the Protobuf definition of the Deployment config (a configuration for distributed training that will be explained later)
Example of dataspec:
Number of records: 1
Number of columns: 3
Number of columns by type:
NUMERICAL: 1 (33%)
CATEGORICAL: 1 (33%)
CATEGORICAL_SET: 1 (33%)
Columns:
NUMERICAL: 6 (40%)
0: "feature_1" NUMERICAL mean:38.5816
CATEGORICAL: 1 (33%)
1: "feature_2" CATEGORICAL has-dict vocab-size:2 zero-ood-items most-frequent:"CAT" 1 (50%)
CATEGORICAL_SET: 1 (33%)
2: "feature_3" CATEGORICAL_SET integerized
Terminology:
nas: Number of non-available (i.e. missing) values.
ood: Out of dictionary.
manually-defined: Attribute which type is manually defined by the user i.e. the type was not automatically inferred.
tokenized: The attribute value is obtained through tokenization.
has-dict: The attribute is attached to a string dictionary e.g. a categorical attribute stored as a string.
vocab-size: Number of unique values.
When stored as a raw file, a dataspec can be read directly. Following is the raw definition of the dataspec printed above.
columns {
name: "attribute_1"
type: NUMERICAL
numerical {
mean: 10
}
}
columns {
name: "attribute_2"
type: CATEGORICAL
categorical {
items {
key: "CAT"
value { index: 1 }
}
items {
key: "COT"
value { index: 2 }
}
}
}
columns {
name: "attribute_3"
type: CATEGORICAL_SET
categorical_set {
is_already_integerized: true
}
}
A dataspecs is already created automatically with the dataspec inference tool
(i.e., infer_dataspec). If some part of the dataspec is incorrectly inferred,
the user can override it with a dataspec guide (see
dataset/data_spec.proto).
For example, integer-looking values are detected as numerical by default. However, integer values representing ENUM values are better suited as categorical. Following is an example of dataspec guide that forces all the attributes with a name starting with the "enum_" prefix to be detected as categorical.
column_guides {
column_name_pattern: "^foo.*" # Regular expression
type: CATEGORICAL # Force this semantic.
}
The following two examples show the inference of a dataspec using the CLI and C++ API:
CLI
infer_dataspec --dataset=csv:/my/dataset.csv --output=dataspec.pbtxt
# Human readable description of the dataspec.
show_dataspec --dataspec=dataspec.pbtxt
C++
using namespace yggdrasil_decision_forests;
dataset::proto::DataSpecification data_spec;
dataset::CreateDataSpec("csv:/my/dataset.csv", false, guide, &data_spec);
# Human description of the dataspec.
std::cout << dataset::PrintHumanReadable(data_spec);
Dataset path and format
The format of a dataset is specified by a prefix and a colon (":") in the
dataset file path. For example, csv:/path/to/my/dataset.csv is a dataset path
to a csv file.
Supported formats are:
csv:: A CSV file.tfrecord+tfe:: A GZIP compressed TensorFlow Record file containing serialized TensorFlow Example protobuffers.
A single dataset can be divided into multiple files. Splitting up the datasets can speed-up dataset reading. As a rule of thumb, individual dataset files should not exceed 100MB. The list of files of a dataset can be specified using sharding, globbing, and comma separation. Following are examples of dataset paths made of multiple files:
csv:/path/to/my/dataset.csv@10csv:/path/to/my/dataset_*.csvcsv:/path/to/my/dataset_1.csv,/path/to/my/dataset_1.csv
In addition, comma separation can be combined with sharding/globbing. For
example: csv:/1/data.csv*,/2/data.csv*.
Learners and Models
YDF operates with the notions of Learners and Models. A learner is a function (in the mathematical sense) that takes a dataset and outputs a model. A model is a function that takes an example, and outputs a prediction.
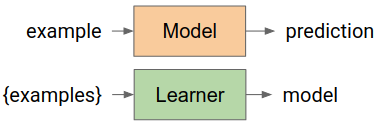
In the model-learner abstraction, a learning algorithm and a set of hyper-parameters is a learner, and a trained model is a model.
A model can be stored in memory or on disk. Information about a model (e.g. input features, label, validation score, learner) can be displayed using the show_model command.
A model also contains metadata. Metadata is not used to compute predictions. Instead, users or tools can query this metadata for analysis (e.g. feature importances, training losses). Model meta-data can be removed using the edit_model tool.
A model can be compiled into an (serving) engine. An engine is a model without metadata and with an internal structure optimized for fast inference. While models are platform agnostic, engines are tied to specific hardware (e.g. require a GPU or AVX512 instructions). Engine inference is orders of magnitude faster than the standard model inference.
The C++ method model.BuildFastEngine() will compile the model into the most
efficient engine on the current hardware. The cli/benchmark_inference tool can
measure the inference speed of a model and the compatible serving engines.
A learner is instantiated from a TrainingConfig proto. The TrainingConfig defines the label, input features, task, learner type, and hyper-parameters for the learning.
The generic hyper-parameters (GHPs) are an alternative representation to the TrainingConfig protobuffer. A GHP is a map of string to values suited for quick configuration, and automated hyper-parameter optimization. GHPs are used by TensorFlow Decision Forests (TF-DF):
The hyper-parameter page lists the learners and their hyper-parameters.
Optionally, a learner can be configured with a deployment specification. A deployment specification defines the computing resources to use during the training. This includes the number of threads or the number of workers (in the case of distributed training). Changing the deployment configuration (or not providing any) can impact the speed of training, but it cannot impact the final model: Two models trained with the same learner, but different deployment specifications are guaranteed to be equal.
By default, learners are configured to be trained locally using 6 threads.
The logging directory of a learner is an optional directory that specifies where the learner exports information about the learning process. This information is learner specific and can be composed of plots, tables and HTML reports, and helps one understand the model. The logging directory of a learner does not have any defined structure as different learners can export different types of information.
The following two examples show the training of a model using the CLI and C++
API (see above how to create the dataspec in the dataspec.pbtxt file):
CLI
cat <<EOF > hparams.pbtxt
task: CLASSIFICATION
label: "income"
learner: "GRADIENT_BOOSTED_TREES"
EOF
train \
--dataset=csv:/my/dataset.csv \
--dataspec=dataspec.pbtxt \
--config=hparams.pbtxt \
--output=/my/model
# Human description of the model.
show_model --model=dataspec.pbtxt
C++
using namespace yggdrasil_decision_forests;
dataset::proto::DataSpecification data_spec = ... // Previous example.
// Configure the training.
model::proto::TrainingConfig config;
config.set_task(model::proto::CLASSIFICATION);
config.set_label("income");
config.set_learner("GRADIENT_BOOSTED_TREES");
// Instantiate the learner.
std::unique_ptr<model::AbstractLearner> learner;
CHECK_OK(model::GetLearner(config, &learner));
// Train
std::unique_ptr<model::AbstractModel> model = learner->Train("csv:/my/dataset.csv", data_spec);
// Export the model to disk.
CHECK_OK(model::SaveModel("/my/model", model.get()));
// Human description of the model.
std::string description;
model->AppendDescriptionAndStatistics(false, &description);
std::cout << description;
To minimize binary sizes and allow the support of custom learning algorithms and models, models, and learners rely on a registration mechanism: Users can restrict or extend the list of supported models and learners with dependency injection.
All the official supported models and learners can be respectively injected with
the dependencies yggdrasil_decision_forests/model:all_models and
yggdrasil_decision_forests/learners:all_learners.
Meta-Learner
Learners can be combined. A learner containing another learner is called a meta-learner. This includes the hyper-parameter optimizer learner, the calibrator learner, the ensembler learner, and the feature selection learner.
Model/Learner Evaluation
Model evaluation is available with the evaluate CLI command or the
model::AbstractModel::Evaluate c++ function).
The following two examples show the evaluation of a model using the CLI and C++ API:
CLI
C++
using namespace yggdrasil_decision_forests;
std::unique_ptr<model::AbstractModel> model = ... // Previous example.
// Loads the dataset.
dataset::VerticalDataset dataset;
CHECK_OK(LoadVerticalDataset("csv:/my/dataset.csv", model->data_spec(), &dataset));
// Evaluate the model.
utils::RandomEngine rnd;
metric::proto::EvaluationResults evaluation = model->Evaluate(dataset, {}, &rnd);
// Export the evaluation in a human-readable format.
std::string text_report;
metric::AppendTextReport(evaluation, &text_report);
std::cout << text_report;
The evaluation of a learner consists of evaluating one or multiple models trained by the learner (e.g. cross-validation; C++ (see model::EvaluateLearner)) . Learner evaluation is more accurate than model evaluation when the dataset is small (e.g. less than 50k examples, or when the learner is unstable).
Model Analysis
The show_model CLI command shows the following information about a model:
- The input features and labels of the model.
- The variable importance of each feature.
- The detailed or summarized structure of the model (e.g. number of nodes).
- The compatible serving engine.
- The training logs.
Alternatively, models can be visualized and analyzed programmatically directly
using the
TensorFlow Decision Forests python inspector
. See the
Inspect and debug
colab for more details. A Yggdrasil model can be converted into a Tensorflow
Decision Forests model using the
tfdf.keras.core.yggdrasil_model_to_keras_model
function. Calling model.summary() shows the same information as the
show_model CLI.
Variable Importances
Variable importance are obtained with CLI (show_model), the C++ API
(model.GetVariableImportance()) and the Python API
(tfdf.inspector.make_inspector(path).variable_importances()).
The available variable importances are:
Model agnostic
MEAN_{INCREASE,DECREASE}_IN_{metric}: Estimated metric change from removing a feature using permutation importance. Depending on the learning algorithm and hyper-parameters, the VIs can be computed with validation, cross-validation or out-of-bag. For example, theMEAN_DECREASE_IN_ACCURACYof a feature is the drop in accuracy (the larger, the most important the feature) caused by shuffling the values of a features. For example,MEAN_DECREASE_IN_AUC_3_VS_OTHERSis the expected drop in AUC when comparing the label class "3" to the others.
Decision Forests specific
-
SUM_SCORE: Sum of the split scores using a specific feature. The larger, the most important. -
NUM_AS_ROOT: Number of root nodes using a specific feature. The larger, the most important. -
NUM_NODES: Number of nodes using a specific feature. The larger, the most important. -
INV_MEAN_MIN_DEPTH: Inverse (i.e. 1/(1+x)) of the average minimum depth of the first occurrence of a feature across all the tree paths. Note:MEAN_MIN_DEPTHwas removed.
Automated Hyper-parameter Tuning
Optimizing the hyperparameters of a learner can improve the quality of a model. Selecting the optimal hyper-parameters can be done manually (see how to improve a model) or using the automated hyper-parameter optimizer (HPO). The HPO automatically selects the best hyper-parameters through a sequence of trial-and-error computations.
Check the example directory for examples of hyper-parameter tuning.
The HPO is a meta-learner with the key HYPERPARAMETER_OPTIMIZER configured
with a sub-learner to optimize. It supports local and distributed training.
Various aspects of the HPO can be configured. The main configuration knobs are.
- The search space: The set of hyper-parameters to optimize.
- The evaluation protocol: How to evaluate a set of hyper-parameters, e.g., self-evaluation, cross-validation.
- The optimizer: The optimization algorithm used to navigate through the search space.
The following example shows the TrainingConfig configuration of the hyper-parameter optimizer for a Gradient boosted trees model.
task: CLASSIFICATION
learner: "HYPERPARAMETER_OPTIMIZER"
label: "income"
[yggdrasil_decision_forests.model.hyperparameters_optimizer_v2.proto
.hyperparameters_optimizer_config] {
retrain_final_model: true # Retraining the model after tuning.
optimizer {
# New hyperparameters values are generated randomly.
optimizer_key: "RANDOM"
[yggdrasil_decision_forests.model.hyperparameters_optimizer_v2.proto
.random] {
num_trials: 25 # 25 Random trials.
}
}
# The base learner to optimize.
base_learner {
# Note: There is no need to configure the label and input features of the
# base learner. The values are automatically copied from the HPO
# configuration.
learner: "GRADIENT_BOOSTED_TREES"
[yggdrasil_decision_forests.model.gradient_boosted_trees.proto
.gradient_boosted_trees_config] {
# The number of trees is fixed.
num_trees: 100
}
}
base_learner_deployment {
# Each candidate learner runs on a single thread.
num_threads: 1
}
# List of hyper-parameters to optimize.
search_space {
fields {
name: "num_candidate_attributes_ratio"
discrete_candidates {
possible_values { real: 1.0 }
possible_values { real: 0.8 }
possible_values { real: 0.6 }
}
}
fields {
name: "use_hessian_gain"
discrete_candidates {
possible_values { categorical: "true" }
possible_values { categorical: "false" }
}
}
fields {
name: "growing_strategy"
discrete_candidates {
possible_values { categorical: "LOCAL" }
possible_values { categorical: "BEST_FIRST_GLOBAL" }
}
children {
parent_discrete_values {
possible_values { categorical: "LOCAL" }
}
name: "max_depth"
discrete_candidates {
possible_values { integer: 4 }
possible_values { integer: 5 }
possible_values { integer: 6 }
possible_values { integer: 7 }
}
}
children {
parent_discrete_values {
possible_values { categorical: "BEST_FIRST_GLOBAL" }
}
name: "max_num_nodes"
discrete_candidates {
possible_values { integer: 16 }
possible_values { integer: 32 }
possible_values { integer: 64 }
possible_values { integer: 128 }
}
}
}
}
}
Distributed Training
Distributed training refers to the training of a model on multiple computers. Distributed training is especially valuable on a large dataset (e.g., >10M examples) or for expensive learning algorithms (e.g., hyper-parameter tuning).
When distributed training is used, the process executing the "train" operation
(i.e. the programming running the learner->Train() function, or the process
running the train CLI command) becomes the manager. The remaining work is
executed by workers. A worker is an instance of the worker binary.
Not all learner support distributed training. For example the
GRADIENT_BOOTED_TREES learner does not support distributed training, however,
DISTRIBUTED_GRADIENT_BOOSTED_TREES does. Some learners (e.g.,
HYPERPARAMETER_OPTIMIZER) supports both distributed and non-distributed
training.
The following learners support distributed training:
-
DISTRIBUTED_GRADIENT_BOOSTED_TREES: Exact distributed training of the gradient boosted tree models. -
HYPERPARAMETER_OPTIMIZER: Automatic optimization of the hyper-parameters of a model.
When training on large datasets, it is recommended for the dataset be separated into multiple files. See the dataset path and format section for more details.
Following is an example of distributed training with the
DISTRIBUTED_GRADIENT_BOOSTED_TREES algorithm.
The training dataset is a set of csv files located in the /remote/dataset/
remote directory. For example:
ls /remote/dataset/
> /remote/dataset/train_0.csv
> /remote/dataset/train_1.csv
> /remote/dataset/train_2.csv
...
> /remote/dataset/train_1000.csv
The dataset path is therefore csv:/remote/dataset/train_*.csv.
Importantly, for the DISTRIBUTED_GRADIENT_BOOSTED_TREES algorithm (different algorithms have different constraints) all the workers should have access to this dataset.
Training configuration is:
train_config.pbtxt
learner: "DISTRIBUTED_GRADIENT_BOOSTED_TREES"
task: CLASSIFICATION
label: "income"
[yggdrasil_decision_forests.model.distributed_gradient_boosted_trees.proto
.distributed_gradient_boosted_trees_config] {
}
Distributed training is enabled by configuring a distribute execution engine
(DEE) and setting the cache_path field in the
DeploymentConfig.
The DEE defines the number of workers and how to reach them (e.g., a list of ip
addresses).
Following is an example of DeploymentConfig configured for distributed training:
deploy_config.pbtxt
# The work_directory is a remote directory accessible to all workers.
cache_path: "/remote/work_directory"
num_threads: 8 # Each worker will run 8 threads.
try_resume_training: true # Allow training to be interrupted and resumed.
distribute {
implementation_key: "GRPC"
[yggdrasil_decision_forests.distribute.proto.grpc] {
socket_addresses {
# Configure the 3 workers.
addresses { ip: "192.168.0.10" port: 8123 }
addresses { ip: "192.168.0.11" port: 8123 }
addresses { ip: "192.168.0.12" port: 8123 }
}
}
}
Before starting the distributed training, you need to start the workers. In this example, we configured 3 workers running on three different computers. On each of those computers, we need to start a worker process with the corresponding port. For example:
# Connect with ssh to 192.168.0.10, and then run:
# Download and extract the worker binary.
wget https://github.com/google/yggdrasil-decision-forests/releases/download/1.0.0/cli_linux.zip
unzip cli_linux.zip
# Start the worker
./grpc_worker_main --port=8123
See yggdrasil_decision_forests/utils/distribute/implementations/grpc/grpc.proto for more details.
Once the worker started, the training can be launched on the manager:
# On the manager.
# Check that all the configuration files are available
ls
> train_config.pbtxt
> deploy_config.pbtxt
# Determine the dataspec of the dataset
# See the "Create dataspec" section in the "CLI / Quick Start" for more details.
./infer_dataspec \
--dataset=csv:/remote/dataset/train_*.csv \
--output=dataspec.pbtxt
# Start the distributed training.
./train \
--dataset=csv:/remote/dataset/train_*.csv \
--dataspec=dataspec.pbtxt \
--config=train_config.pbtxt\
--deployment=deploy_config.pbtxt\
--output=model
This example demonstrates distributed training by running all the workers on the same machine. This is effectively not distributed training, but this is an easy way to test it.
The example above used the GRPC DEE. Different DEE determines how distributed
computation primitives are executed. The choice of the DEE does not impact the
effective trained model.
For example, TensorFlow Decision Forests uses the [TF_DIST]](https://github.com/tensorflow/decision-forests/blob/main/tensorflow_decision_forests/tensorflow/distribute/tf_distribution.proto) DEE which can run distributed computation of TensorFlow Parameter Servers. The TF_DIST DEE supports both socket addresses and with the TF_CONFIG configuration
The GRPC distribute engine is recommended for the CLI or C++ APIs. When using the TensorFlow Decision Forests interface, both the GRPC and TF_DIST are viable options (with pro and cons).
Feature Engineering
Improving the features is important for the model quality. For the sake of simplicity (YDF does only one thing) and execution efficiency (direct code execution is often much faster than intermediate representation execution), YDF core library does not contain customizable processing components with the exception of text tokenization.
Registered classes
Yggdrasil decision forest's source code is divided into modules. Different modules implement different functions such as learning algorithms, models, support for dataset formats, etc.
Modules are controlled by a registration mechanism through Bazel dependency
rules: To enable a module, a dependency to this module should be added to
any of the cc_library or cc_binary of the code. Note that adding modules
also increases the size of the binary.
For simplicity, CLI tools in yggdrasil_decision_forests/cli are compiled with
all the available modules.
When using the C++ API, the dependency to modules should be added manually. For
example, to support the training of RANDOM_FOREST models, the binary/library
needs to depend on the "yggdrasil_decision_forests/learner/random_forest" rule.
An error of the type "No class registered with key..." or "Unknown item ... in class pool" indicates that a dependency to a required module is missing.
Yggdrasil decision forest also defines module groups that contain all the
modules of a certain type. For example, the rule
yggdrasil_decision_forests/learner:all_learners injects all the available
learning algorithms (including the :random_forest one mentioned above).
Following is the list the available module path and registration keys.
Learning algorithms
- learner/cart CART
- learner/distributed_gradient_boosted_trees DISTRIBUTED_GRADIENT_BOOSTED_TREES
- learner/gradient_boosted_trees GRADIENT_BOOSTED_TREES
- learner/random_forest RANDOM_FOREST
- learner/hyperparameters_optimizer HYPERPARAMETER_OPTIMIZER
Models
- model/gradient_boosted_trees GRADIENT_BOOSTED_TREES
- model/random_forest RANDOM_FOREST
Inference engines
- serving/decision_forest:register_engines
Dataset IO
- dataset:csv_example_reader FORMAT_CSV (reading only)
- dataset:csv_example_writer FORMAT_CSV (writing only)
- dataset:tf_example_io_tfrecord FORMAT_TFE_TFRECORD
- dataset:capacitor_example_reader FORMAT_CAPACITOR
- dataset:tf_example_io_recordio FORMAT_TFE_RECORDIO
- dataset:tf_example_io_sstable FORMAT_TFE_SSTABLE
Distributed computation backends
- utils/distribute/implementations/multi_thread MULTI_THREAD
- utils/distribute/implementations/grpc GRPC
- tensorflow_decision_forests/tensorflow/distribute:tf_distribution TF_DIST
Losses for the gradient boosted trees learner
- learner/gradient_boosted_trees/loss/loss_imp_*
Advanced features
Most of the YDF advanced documentation is written in the .h headers (for the
library) and .cc headers (for CLI tools). ~~~